8. Processing multidimensional data¶
This open-access textbook is, and will remain, freely available for everyone’s enjoyment (also in PDF; a paper copy can also be ordered). It is a non-profit project. Although available online, it is a whole course, and should be read from the beginning to the end. Refer to the Preface for general introductory remarks. Any bug/typo reports/fixes are appreciated. Make sure to check out Deep R Programming [36] too.
8.1. Extending vectorised operations to matrices¶
The vector operations from Chapter 5 are brilliant examples of the write less, do more principle in practice. Let’s see how are they extended to matrices.
8.1.1. Vectorised mathematical functions¶
Applying vectorised functions such as numpy.round, numpy.log, and numpy.exp returns an array of the same shape, with all elements transformed accordingly:
A = np.array([
[0.2, 0.6, 0.4, 0.4],
[0.0, 0.2, 0.4, 0.7],
[0.8, 0.8, 0.2, 0.1]
]) # example matrix
For instance, to take the square of every element, we can call:
np.square(A)
## array([[0.04, 0.36, 0.16, 0.16],
## [0. , 0.04, 0.16, 0.49],
## [0.64, 0.64, 0.04, 0.01]])
8.1.2. Componentwise aggregation¶
Unidimensional aggregation functions (e.g., numpy.mean, numpy.quantile) can be applied to summarise:
all data into a single number (
axis=None, being the default),data in each column (
axis=0), as well asdata in each row (
axis=1).
Here are the corresponding examples:
np.mean(A)
## 0.39999999999999997
np.mean(A, axis=0)
## array([0.33333333, 0.53333333, 0.33333333, 0.4 ])
np.mean(A, axis=1)
## array([0.4 , 0.325, 0.475])
Important
Let’s stress that axis=1 does not mean that we get the column means
(even though columns constitute the second axis, and we count starting at 0).
It denotes the axis along which the matrix is sliced.
Sadly, even yours truly sometimes does not get it right.
Given the nhanes_adult_female_bmx_2020
dataset, compute the mean, standard deviation, the minimum,
and the maximum of each body measure.
We will get back to the topic of the aggregation of multidimensional data in Section 8.4.
8.1.3. Arithmetic, logical, and relational operations¶
Recall that for vectors, binary operators such as `+`, `*`, `==`, `<=`, and `&` as well as similar elementwise functions (e.g., numpy.minimum) can be applied if both inputs are of the same length. For example:
np.array([1, 10, 100, 1000]) * np.array([7, -6, 2, 8]) # elementwisely
## array([ 7, -60, 200, 8000])
Alternatively, one input can be a scalar:
np.array([1, 10, 100, 1000]) * -3
## array([ -3, -30, -300, -3000])
More generally, a set of rules referred to in the numpy manual as broadcasting describes how this package handles arrays of different shapes.
Important
Generally, for two matrices, their column/row counts must match or be equal to 1. Also, if one operand is a one-dimensional array, it will be promoted to a row vector.
Let’s explore all the possible scenarios.
8.1.3.1. Matrix vs scalar¶
If one operand is a scalar, then it is going to be propagated over all matrix elements. For example:
(-1)*A
## array([[-0.2, -0.6, -0.4, -0.4],
## [-0. , -0.2, -0.4, -0.7],
## [-0.8, -0.8, -0.2, -0.1]])
It changed the sign of every element, which is, mathematically, an instance of multiplying a matrix \(\mathbf{X}\) by a scalar \(c\):
Furthermore, we can take the square[1] of each element:
A**2
## array([[0.04, 0.36, 0.16, 0.16],
## [0. , 0.04, 0.16, 0.49],
## [0.64, 0.64, 0.04, 0.01]])
or compare each element to 0.25.
A >= 0.25
## array([[False, True, True, True],
## [False, False, True, True],
## [ True, True, False, False]])
8.1.3.2. Matrix vs matrix¶
For two matrices of identical sizes, we act on the corresponding elements:
B = np.tri(A.shape[0], A.shape[1]) # just an example
B # a lower triangular 0-1 matrix
## array([[1., 0., 0., 0.],
## [1., 1., 0., 0.],
## [1., 1., 1., 0.]])
And now:
A * B
## array([[0.2, 0. , 0. , 0. ],
## [0. , 0.2, 0. , 0. ],
## [0.8, 0.8, 0.2, 0. ]])
multiplies each \(a_{i,j}\) by the corresponding \(b_{i,j}\).
This behaviour extends upon the idea from algebra that given \(\mathbf{A}\) and \(\mathbf{B}\) with \(n\) rows and \(m\) columns each, the result of \(+\) is:
Thanks to the matrix-matrix and matrix-scalar operations, we can perform various tests on a per-element basis, e.g.,
(A >= 0.25) & (A <= 0.75) # logical matrix & logical matrix
## array([[False, True, True, True],
## [False, False, True, True],
## [False, False, False, False]])
(*) Figure 8.1 depicts a (filled) contour plot of Himmelblau’s function, \(f(x,y)=(x^{2}+y-11)^{2}+(x+y^{2}-7)^{2}\), for \(x\in[-5, 5]\) and \(y\in[-4, 4]\). To draw it, we probe 250 points from these two intervals, and call numpy.meshgrid to generate two matrices, both of shape 250 by 250, giving the x- and y-coordinates of all the points on the corresponding two-dimensional grid. Thanks to this, we are able to use vectorised mathematical operations to compute the values of \(f\) thereon.
x = np.linspace(-5, 5, 250)
y = np.linspace(-4, 4, 250)
xg, yg = np.meshgrid(x, y)
z = (xg**2 + yg - 11)**2 + (xg + yg**2 - 7)**2
plt.contourf(x, y, z, levels=20)
CS = plt.contour(x, y, z, levels=[1, 5, 10, 20, 50, 100, 150, 200, 250])
plt.clabel(CS, colors="black")
plt.show()
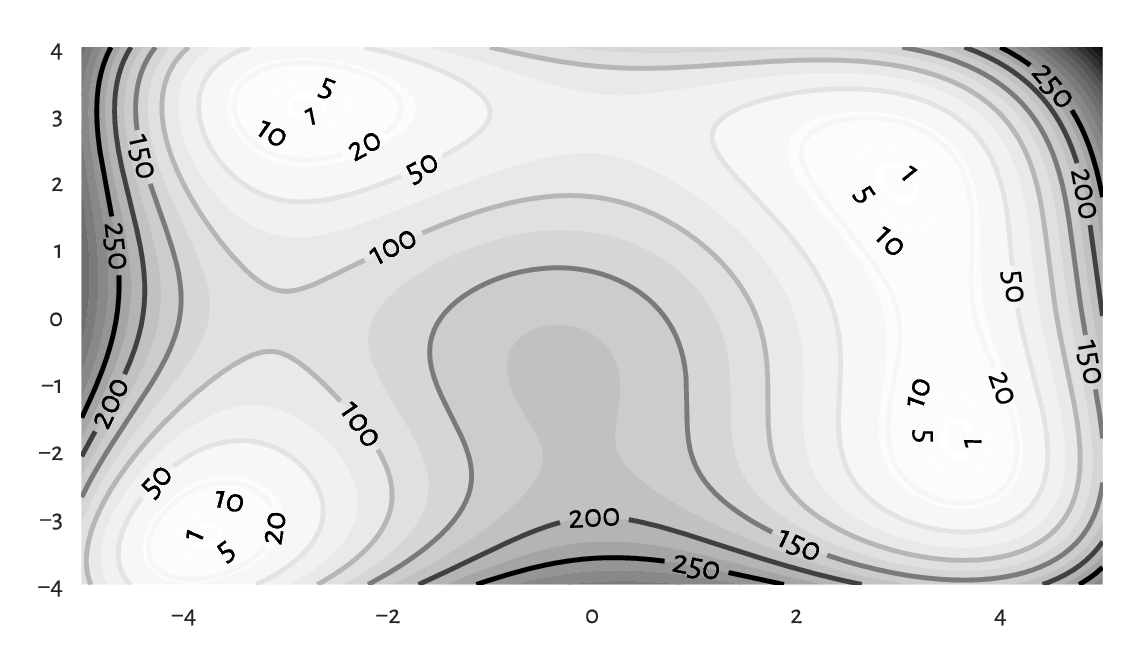
Figure 8.1 An example filled contour plot with additional labelled contour lines.¶
To understand the result generated by numpy.meshgrid, let’s inspect its output for a smaller number of probe points:
x = np.linspace(-5, 5, 3)
y = np.linspace(-4, 4, 5)
xg, yg = np.meshgrid(x, y)
xg
## array([[-5., 0., 5.],
## [-5., 0., 5.],
## [-5., 0., 5.],
## [-5., 0., 5.],
## [-5., 0., 5.]])
Here, each column consists of the same values.
yg
## array([[-4., -4., -4.],
## [-2., -2., -2.],
## [ 0., 0., 0.],
## [ 2., 2., 2.],
## [ 4., 4., 4.]])
In this case, each row is constant. Therefore, calling:
(xg**2 + yg - 11)**2 + (xg + yg**2 - 7)**2
## array([[116., 306., 296.],
## [208., 178., 148.],
## [340., 170., 200.],
## [320., 90., 260.],
## [340., 130., 520.]])
gives a matrix \(\mathbf{Z}\) such that \(z_{i,j}\) is generated by considering
the \(i\)-th element in y and the \(j\)-th item in x, which is exactly what
we desired. We will provide an alternative implementation in
Example 8.5.
8.1.3.3. Matrix vs any vector¶
An n×m matrix can also be combined with an n×1 column vector:
A * np.array([1, 10, 100]).reshape(-1, 1)
## array([[ 0.2, 0.6, 0.4, 0.4],
## [ 0. , 2. , 4. , 7. ],
## [80. , 80. , 20. , 10. ]])
It propagated the column vector over all columns (left to right). Similarly, combining a matrix with a 1×m row vector recycles the latter over all rows (top to bottom).
A + np.array([1, 2, 3, 4]).reshape(1, -1)
## array([[1.2, 2.6, 3.4, 4.4],
## [1. , 2.2, 3.4, 4.7],
## [1.8, 2.8, 3.2, 4.1]])
If one operand is a one-dimensional array or a list of length \(m\), it will be treated as a row vector. For example, here is an instance of centring of each column:
np.round(A - np.mean(A, axis=0), 3) # matrix - vector
## array([[-0.133, 0.067, 0.067, -0. ],
## [-0.333, -0.333, 0.067, 0.3 ],
## [ 0.467, 0.267, -0.133, -0.3 ]])
An explicit .reshape(1, -1) was not necessary.
Mathematically, although it is not necessarily a standard notation, we will allow adding and subtracting row vectors from matrices of compatible sizes:
This corresponds to shifting (translating) every row in the matrix.
In the nhanes_adult_female_bmx_2020
dataset, standardise, normalise, and min-max scale every column
(compare Section 5.3.2).
A single line of code will suffice in each case.
8.1.3.4. Row vector vs column vector (*)¶
A row vector combined with a column vector results in an operation’s being performed on each combination of all pairs of elements in the two arrays (i.e., the cross-product; not just the corresponding pairs).
np.arange(1, 8).reshape(1, -1) * np.array([1, 10, 100]).reshape(-1, 1)
## array([[ 1, 2, 3, 4, 5, 6, 7],
## [ 10, 20, 30, 40, 50, 60, 70],
## [100, 200, 300, 400, 500, 600, 700]])
Check out that numpy.nonzero relies on similar shape broadcasting rules as the binary operators we discussed here, but not with respect to all three arguments.
(*) Himmelblau’s function in Example 8.2 is defined by means of arithmetic operators only, and they all rely on the kind of shape broadcasting that we discuss in this section. Consequently, calling numpy.meshgrid to evaluate \(f\) on a point grid was not really necessary:
x = np.linspace(-5, 5, 3)
y = np.linspace(-4, 4, 5)
xg = x.reshape(1, -1)
yg = y.reshape(-1, 1)
(xg**2 + yg - 11)**2 + (xg + yg**2 - 7)**2
## array([[116., 306., 296.],
## [208., 178., 148.],
## [340., 170., 200.],
## [320., 90., 260.],
## [340., 130., 520.]])
See also the sparse parameter in numpy.meshgrid,
and Section 12.3.1 where this function turns out useful
after all.
8.1.4. Other row and column transforms (*)¶
Some functions discussed in the previous part of this course
are equipped with the axis argument, which
supports processing each row or column independently.
For example, to compute the ranks of elements in each column, we can call:
scipy.stats.rankdata(A, axis=0) # columnwisely (along the rows)
## array([[2. , 2. , 2.5, 2. ],
## [1. , 1. , 2.5, 3. ],
## [3. , 3. , 1. , 1. ]])
Some functions have the default argument axis=-1
meaning that they are applied along the last[2] axis
(i.e., columns in the matrix case):
np.diff(A) # means axis=1 in this context (along the columns)
## array([[ 0.4, -0.2, 0. ],
## [ 0.2, 0.2, 0.3],
## [ 0. , -0.6, -0.1]])
Compare the foregoing to the iterated differences in each column separately (along the rows):
np.diff(A, axis=0)
## array([[-0.2, -0.4, 0. , 0.3],
## [ 0.8, 0.6, -0.2, -0.6]])
If a vectorised function in not equipped with the axis argument,
we can propagate it over all the rows or columns by calling
numpy.apply_along_axis. For instance, here is another
(did you solve Exercise 8.3?)
way to compute the z-scores in each matrix column:
def standardise(x):
return (x-np.mean(x))/np.std(x)
np.round(np.apply_along_axis(standardise, 0, A), 2) # round for readability
## array([[-0.39, 0.27, 0.71, -0. ],
## [-0.98, -1.34, 0.71, 1.22],
## [ 1.37, 1.07, -1.41, -1.22]])
But, of course, we prefer
(x-np.mean(x, axis=0))/np.std(x, axis=0).
Note
(*) Matrices are iterable (in the sense of Section 3.4), but in an interesting way. Namely, an iterator traverses through each row in a matrix. Writing:
r1, r2, r3 = A # A has three rows
creates three variables, each representing a separate row
in A, the second of which is:
r2
## array([0. , 0.2, 0.4, 0.7])
8.2. Indexing matrices¶
Recall that for unidimensional arrays, we have four possible
indexer choices (i.e., when performing filtering like x[i]):
scalar (extracts a single element),
slice (selects a regular subsequence, e.g., every second element or the first six items; returns a view of existing data: it does not make an independent copy of the subsetted elements),
integer vector (selects the elements at given indexes),
logical vector (selects the elements that correspond to
Truein the indexer).
Matrices are two-dimensional arrays. Subsetting thus requires two indexes.
By writing A[i, j], we select rows given by i and columns given by j.
Both i and j can be one of the four aforementioned types,
so we have at ten different cases to consider
(skipping the symmetric ones).
Important
Generally:
each scalar index reduces the dimensionality of the subsetted object by 1;
slice-slice and slice-scalar indexing returns a view of the existing array, so we need to be careful when modifying the resulting object;
usually, indexing returns a submatrix (subblock), which is a combination of elements at given rows and columns;
indexing with two integer or logical vectors is performed elementwisely, and should be avoided if find the rules of shape broadcasting too complicated.
8.2.1. Slice-based indexing¶
Our favourite example matrix again:
A = np.array([
[0.2, 0.6, 0.4, 0.4],
[0.0, 0.2, 0.4, 0.7],
[0.8, 0.8, 0.2, 0.1]
])
Indexing based on two slices selects a submatrix:
A[::2, 3:] # every second row, skip the first three columns
## array([[0.4],
## [0.1]])
An empty slice selects all elements on the corresponding axis:
A[:, ::-1] # all rows, reversed columns
## array([[0.4, 0.4, 0.6, 0.2],
## [0.7, 0.4, 0.2, 0. ],
## [0.1, 0.2, 0.8, 0.8]])
Let’s stress that the result is always in the form of a matrix.
8.2.2. Scalar-based indexing¶
Indexing by a scalar selects a given row or column, reducing the dimensionality of the output object:
A[:, 3] # one scalar: from two to one dimensions
## array([0.4, 0.7, 0.1])
It selected the fourth column and gave a flat vector (we can always use the reshape method to convert the resulting object back to a matrix). Furthermore:
A[0, -1] # two scalars: from two to zero dimensions
## 0.4
It yielded the element (scalar) in the first row and the last column.
8.2.3. Mixed logical/integer vector and scalar/slice indexers¶
A logical and integer vector-like object can also be employed for element selection. If the other indexer is a slice or a scalar, the result is quite predictable. For instance:
A[ [0, -1, 0], ::-1 ]
## array([[0.4, 0.4, 0.6, 0.2],
## [0.1, 0.2, 0.8, 0.8],
## [0.4, 0.4, 0.6, 0.2]])
It selected the first, the last, and the first row again. Then, it reversed the order of columns.
A[ A[:, 0] > 0.1, : ]
## array([[0.2, 0.6, 0.4, 0.4],
## [0.8, 0.8, 0.2, 0.1]])
It chose the rows from A where the values in the first column of A
are greater than 0.1.
A[np.mean(A, axis=1) > 0.35, : ]
## array([[0.2, 0.6, 0.4, 0.4],
## [0.8, 0.8, 0.2, 0.1]])
It fetched the rows whose mean is greater than 0.35.
A[np.argsort(A[:, 0]), : ]
## array([[0. , 0.2, 0.4, 0.7],
## [0.2, 0.6, 0.4, 0.4],
## [0.8, 0.8, 0.2, 0.1]])
It ordered the matrix with respect to the values in the first column (all rows permuted in the same way, together).
In the
nhanes_adult_female_bmx_2020 dataset,
select all the participants whose heights are within their
mean ± 2 standard deviations.
8.2.4. Two vectors as indexers (*)¶
Indexing based on two logical or integer vectors is a tad more horrible, as in this case not only some form of shape broadcasting comes into play but also all the headache-inducing exceptions listed in the perhaps not the most clearly written Advanced Indexing section of the numpy manual. Cheer up, though: Section 10.5 points out that indexing in pandas is even more troublesome.
For the sake of our maintaining sanity, in practice, it is best to be extra careful when using two vector indexers and stick only to the scenarios discussed beneath. First, with two flat integer indexers, we pick elementwisely:
A[ [0, -1, 0, 2, 0], [1, 2, 0, 2, 1] ]
## array([0.6, 0.2, 0.2, 0.2, 0.6])
It yielded A[0, 1], A[-1, 2], A[0, 0], A[2, 2], and A[0, 1].
Second, to select a submatrix (a subblock) using integer indexes, it is best to make sure that the first indexer is a column vector, and the second one is a row vector (or some objects like these, e.g., compatible lists of lists).
A[ [[0], [-1]], [[1, 3]] ] # column vector-like list, row vector-like list
## array([[0.6, 0.4],
## [0.8, 0.1]])
Third, if indexing involves logical vectors, it is best to convert them to integer ones first (e.g., by calling numpy.flatnonzero).
A[ np.flatnonzero(np.mean(A, axis=1) > 0.35).reshape(-1, 1), [[0, 2, 3, 0]] ]
## array([[0.2, 0.4, 0.4, 0.2],
## [0.8, 0.2, 0.1, 0.8]])
The necessary reshaping can be outsourced to numpy.ix_ function:
A[ np.ix_( np.mean(A, axis=1) > 0.35, [0, 2, 3, 0] ) ] # np.ix_(rows, cols)
## array([[0.2, 0.4, 0.4, 0.2],
## [0.8, 0.2, 0.1, 0.8]])
Alternatively, we can always apply indexing twice:
A[np.mean(A, axis=1) > 0.45, :][:, [0, 2, 3, 0]]
## array([[0.8, 0.2, 0.1, 0.8]])
This is only a mild inconvenience. We will be forced to apply such double indexing anyway in pandas whenever selecting rows by position and columns by name is required; see Section 10.5.
Note
(*) Interestingly, we can also index a vector using an integer matrix. This is like subsetting using a list of integer indexes, but the output’s shape matches that of the indexer:
u = np.array(["a", "b", "c"])
V = np.array([ [0, 1], [1, 0], [2, 1] ])
u[V] # like u[V.ravel()].reshape(V.shape)
## array([['a', 'b'],
## ['b', 'a'],
## ['c', 'b']], dtype='<U1')
8.2.5. Views of existing arrays (*)¶
Only the indexing involving two slices or a slice and a scalar returns a view on an existing array. For example:
B = A[:, ::2]
B
## array([[0.2, 0.4],
## [0. , 0.4],
## [0.8, 0.2]])
Now B and A share memory. By modifying B in place, e.g.:
B *= -1
the changes will be visible in A as well:
A
## array([[-0.2, 0.6, -0.4, 0.4],
## [-0. , 0.2, -0.4, 0.7],
## [-0.8, 0.8, -0.2, 0.1]])
This is time and memory efficient, but might lead to some unexpected results if we are being rather absent-minded. The readers have been warned.
In all other cases, we get a copy of the subsetted array.
8.2.6. Adding and modifying rows and columns¶
With slice- and scalar-based indexers, rows, columns, or individual elements can be replaced by new content in a natural way:
A[:, 0] = A[:, 0]**2
With numpy arrays, however, new rows or columns cannot be added via the index operator. Instead, the whole array needs to be created from scratch using, e.g., one of the functions discussed in Section 7.1.4. For example:
A = np.column_stack((A, np.sqrt(A[:, 0])))
A
## array([[ 0.04, 0.6 , -0.4 , 0.4 , 0.2 ],
## [ 0. , 0.2 , -0.4 , 0.7 , 0. ],
## [ 0.64, 0.8 , -0.2 , 0.1 , 0.8 ]])
8.3. Matrix multiplication, dot products, and Euclidean norm (*)¶
Matrix algebra is at the core of all the methods used in data analysis with the matrix multiplication being often the most crucial operation (e.g., [22, 42]). Given \(\mathbf{A}\in\mathbb{R}^{n\times p}\) and \(\mathbf{B}\in\mathbb{R}^{p\times m}\), their multiply is a matrix \(\mathbf{C}=\mathbf{A}\mathbf{B}\in\mathbb{R}^{n\times m}\) such that \(c_{i,j}\) is the sum of the \(i\)-th row in \(\mathbf{A}\) and the \(j\)-th column in \(\mathbf{B}\) multiplied elementwisely:
for \(i=1,\dots,n\) and \(j=1,\dots,m\). For example:
A = np.array([
[1, 0, 1],
[2, 2, 1],
[3, 2, 0],
[1, 2, 3],
[0, 0, 1],
])
B = np.array([
[1, 0, 0, 0],
[0, 4, 1, 3],
[2, 0, 3, 1],
])
And now:
C = A @ B # or: A.dot(B)
C
## array([[ 3, 0, 3, 1],
## [ 4, 8, 5, 7],
## [ 3, 8, 2, 6],
## [ 7, 8, 11, 9],
## [ 2, 0, 3, 1]])
Mathematically, we can denote it by:
For example, the element in the fourth row and the third column, \(c_{4,3}\) takes the fourth row in the left matrix \(\mathbf{a}_{4,\cdot}=[1\ 2\ 3]\) and the third column in the right matrix \(\mathbf{b}_{\cdot,3}=[0\ 1\ 3]^T\) (emphasised with bold), multiplies the corresponding elements, and computes their sum, i.e., \(c_{4,3}=1\cdot 0 + 2\cdot 1 + 3\cdot 3 = 11\).
Important
Matrix multiplication can only be performed on two matrices of compatible sizes: the number of columns in the left matrix must match the number of rows in the right operand.
Another example:
A = np.array([
[1, 2],
[3, 4]
])
I = np.array([ # np.eye(2)
[1, 0],
[0, 1]
])
A @ I # or A.dot(I)
## array([[1, 2],
## [3, 4]])
We matrix-multiplied \(\mathbf{A}\) by the identity matrix \(\mathbf{I}\), which is the neutral element of the said operation. This is why the result is identical to \(\mathbf{A}\).
Important
In most textbooks, just like in this one, \(\mathbf{A}\mathbf{B}\) always denotes the matrix multiplication. This is a very different operation from the elementwise multiplication.
Compare the above to:
A * I # elementwise multiplication (the Hadamard product)
## array([[1, 0],
## [0, 4]])
(*) Show that \((\mathbf{A}\mathbf{B})^T=\mathbf{B}^T \mathbf{A}^T\). Also notice that, typically, matrix multiplication is not commutative, i.e., \(\mathbf{A}\mathbf{B}\neq\mathbf{B}\mathbf{A}\).
Note
By definition, matrix multiplication is convenient for denoting sums of products of corresponding elements in many pairs of vectors, which we refer to as dot products. More formally, given two vectors \(\boldsymbol{x}, \boldsymbol{y} \in \mathbb{R}^p\), their dot (or scalar) product is:
In matrix multiplication terms, if \(\mathbf{x}\) is a row vector and \(\mathbf{y}^T\) is a column vector, then the above can be written as \(\mathbf{x} \mathbf{y}^T\). The result is a single number.
In particular, the dot product of a vector and itself:
is the square of the Euclidean norm of \(\boldsymbol{x}\) (simply the sum of squares), which is used to measure the magnitude of a vector (Section 5.3.2.3):
It is worth pointing out that the Euclidean norm fulfils (amongst others) the condition that \(\|\boldsymbol{x}\|=0\) if and only if \(\boldsymbol{x}=\mathbf{0}=(0,0,\dots,0)\). The same naturally holds for its square.
Show that \(\mathbf{A}^T \mathbf{A}\) gives the matrix that consists of the dot products of all the pairs of columns in \(\mathbf{A}\) and \(\mathbf{A} \mathbf{A}^T\) stores the dot products of all the pairs of rows.
Section 9.3.2 will note that matrix multiplication can be used as a way to express certain geometrical transformations of points in a dataset, e.g., scaling and rotating. Also, Section 9.3.3 briefly discusses the concept of the inverse of a matrix. Furthermore, Section 9.3.4 introduces its singular value decomposition.
8.5. Exercises¶
Does numpy.mean(A, axis=0)
compute the rowwise or columnwise means of A?
How does shape broadcasting work? List the most common pairs of shape cases when performing arithmetic operations like addition or multiplication.
What are the possible ways to index a matrix?
Which kinds of matrix indexers return a view of an existing array?
(*) How can we select a submatrix comprised of the first and the last row and the first and the last column?
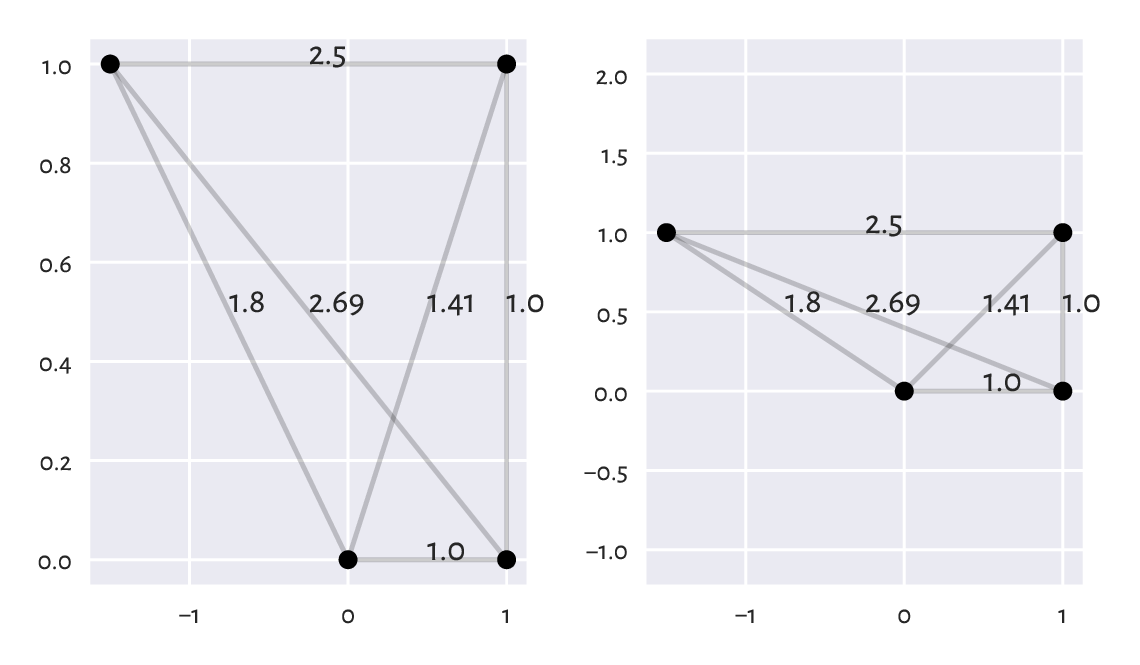
Why appropriate data preprocessing is required when computing the Euclidean distance between the points in a matrix?
What is the relationship between the dot product, the Euclidean norm, and the Euclidean distance?
What is a centroid? How is it defined by means of the Euclidean distance between the points in a dataset?
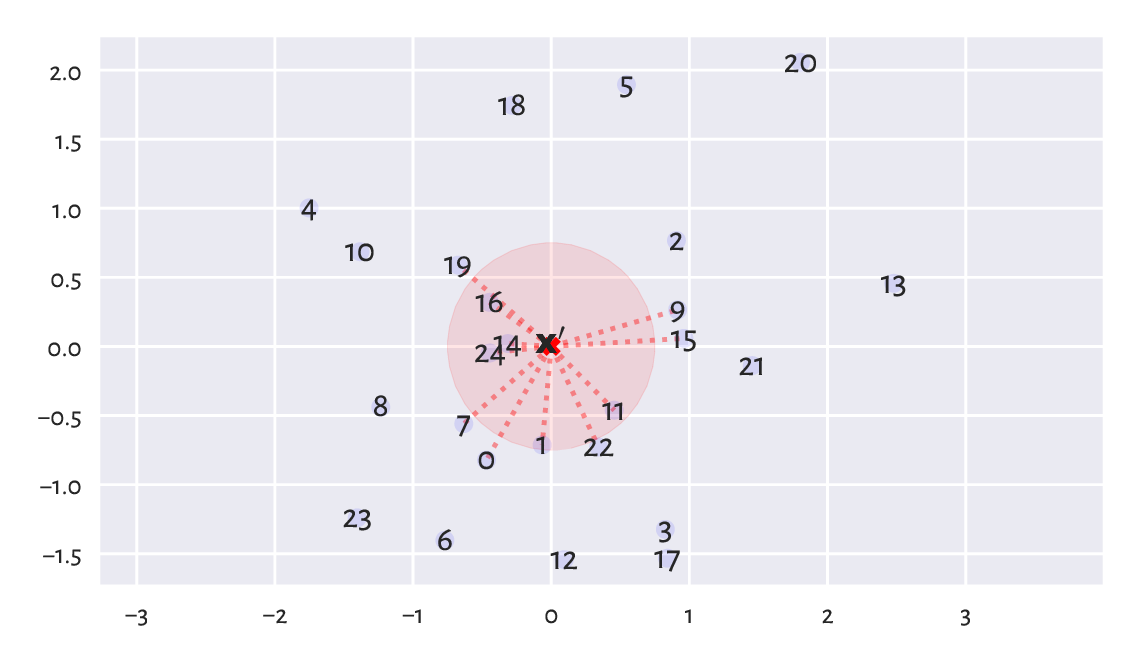
What is the difference between the fixed-radius and the few nearest neighbour search?
(*) When \(K\)-d trees or other spatial search data structures might be better than a brute-force search based on scipy.spatial.distance.cdist?
(**) See what kind of vector and matrix processing capabilities are available in the following packages: TensorFlow, PyTorch, Theano, and tinygrad. Are their APIs similar to that of numpy?